Microsoft ends support for Internet Explorer on June 16, 2022.
We recommend using one of the browsers listed below.
- Microsoft Edge(Latest version)
- Mozilla Firefox(Latest version)
- Google Chrome(Latest version)
- Apple Safari(Latest version)
Please contact your browser provider for download and installation instructions.
February 4, 2025
NTT Corporation
Meiji University
First in the World to Control the Long-Term Survival of Microorganisms in Soil
Establishment of fundamental technologies for reducing greenhouse gas emissions from soil
News Highlights:
- We have developed a foundational technology to control the long-term survival of microorganisms in soil, contributing to a reduction in environmental impact
- By targeting all transcription factors1 in a single bacterium (E. coli), we comprehensively identified the genes required for the long-term survival of bacteria in soil
- By modifying the viability of microorganisms in soil, this technology is expected to reduce greenhouse gas emissions and minimize the use of chemical fertilizers by optimizing material circulation in the soil
TOKYO - February 4, 2025 - NTT Corporation (Headquarters: Chiyoda Ward, Tokyo; Representative Member of the Board and President: Akira Shimada; hereinafter "NTT") and Associate Professor Tomohiro Shimada of the School of Agriculture, Meiji University (Headquarters: Chiyoda Ward, Tokyo; President: Masao Ueno) have successfully identified genes that determine the viability of microorganisms in soil. Using Escherichia coli (E. coli)2 as a model microorganism, we have identified multiple genes that contribute to the long-term viability of microorganisms in soil for the first time in the world. This breakthrough is expected to serve as a foundational technology for reducing environmental impact—such as decreasing greenhouse gas emissions from soil and lowering the use of chemical fertilizers by optimizing material circulation in soil.
The findings were published in the British scientific journal Scientific Reports on February 4, 2025.
Background
According to the Intergovernmental Panel on Climate Change (IPCC) Sixth Assessment Report (2021)3, "It is unequivocal that human influence has warmed the atmosphere, ocean and land." and urgent action is needed to mitigate global warming. Emissions of carbon dioxide (CO2), a key greenhouse gas, from land, including soil are reported to be about 12 times higher than those from human activities (Figure 1). Nitrous oxide (N2O), which has a greenhouse effect approximately 290 times4 greater than CO2, is produced by the addition of excessive chemical fertilizers and the activity of soil microorganisms (Figure 2). In addition, nutrients such as nitrogen that are not absorbed by plants flow into rivers and other external environments, causing damage to ecosystems and placing further strain on the environment. This highlights the need for technology that can appropriately control the activity of microorganisms in soil and reduce environmental burdens.
Until now, the main method for controlling the activity of microorganisms in soil have focused on changing soil's physical properties—such as hardness, water retention, and air permeability—or altering chemical factors, such as soil pH and the quantity and type of nutrients. However, as these methods can only indirectly affect the microbial flora, they cannot reliably increase or decrease the amount of specific microbial species. For example, efforts to increase the microorganisms that convert N2O have often failed to specifically target these species, limiting the ability to reduce greenhouse gas emissions effectively.
To address this challenge, it has become necessary to identify the genes that determine the viability of microorganisms in soil and develop technologies that can individually control the viability of specific microbial species. Building on this concept, NTT and Meiji University have been conducting joint research to identify the genes of E. coli as a model microorganism.
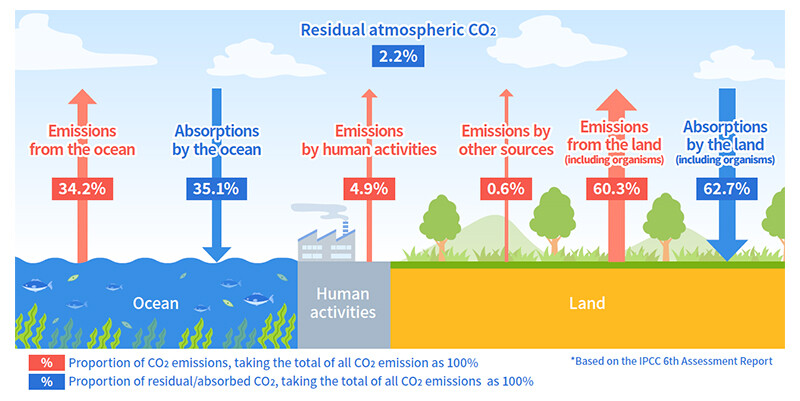 Figure 1 Carbon Dioxide (CO2) Cycle on Earth
Figure 1 Carbon Dioxide (CO2) Cycle on Earth
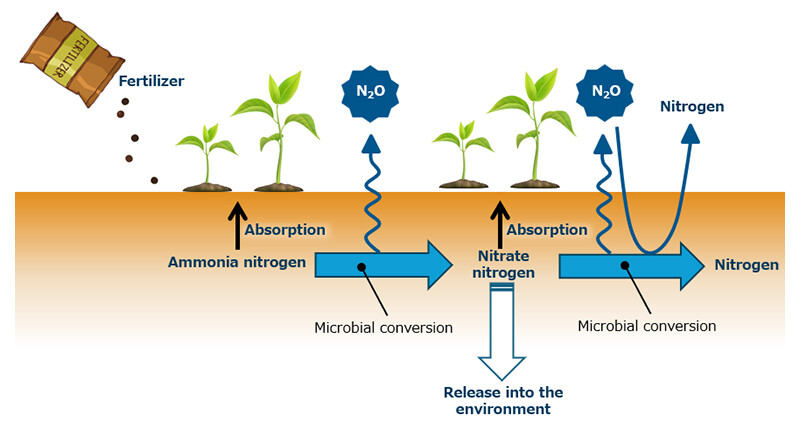 Figure 2 Overview of the Conversion of Nitrogen Compounds by Microorganisms in Soil.
Figure 2 Overview of the Conversion of Nitrogen Compounds by Microorganisms in Soil.
Soil Microorganisms Convert Ammonia Nitrogen to Nitrate Nitrogen, Nitrate Nitrogen to Nitrogen, And Nitrous Oxide (N2O) to Nitrogen.
Key points of technology and outline of experiments
(1) Method for determining the viability of E. coli in soil
The long-term viability of microorganisms in soil is not well understood, particularly regarding the genes involved. To address this, we utilized E. coli, a model microorganism with extensive genetic data, providing advanced insights into gene function.
To evaluate the long-term viability of E. coli in soil, we developed a measurement method. Approximately 20 million E. coli wild-type cells were mixed with 1 g of soil and placed in a constant temperature and humidity chamber set to 25℃ and 60% humidity. The standard method for assessing viable cells in soil involves quantifying genomic DNA via PCR5. However, this method cannot differentiate between living and dead cells. In this study, we determined the number of truly viable cells by counting the colonies6 formed by viable cells on agar plates7 (Figure 3A) at specific time points: days 0, 3, 7, 21, and 42. The results showed that viability decreased over time, with colony formation rates of 7.4% (day 3), 4.3% (day 7), 1.1% (day 21), and 0.27% (day 42), compared to 100% survival on day 0 (Figure 3B).
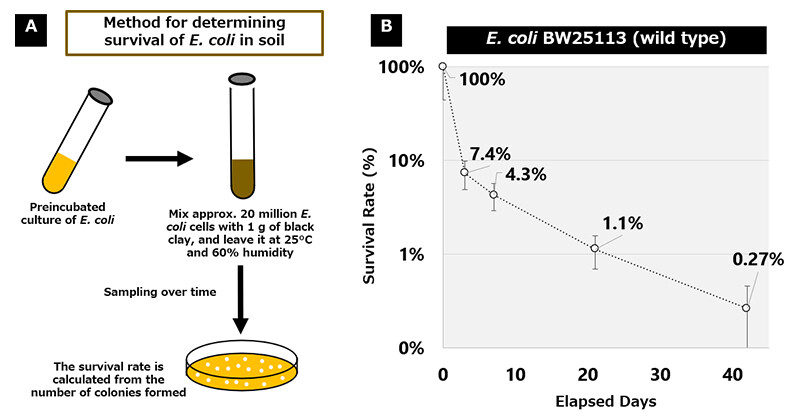 Figure 3 Experimental System for Measuring the Survival Rate of E. coli in Soil (A) and Transition of the Survival Rate of the Wild Type Strain of E. coli in Soil (B)
Figure 3 Experimental System for Measuring the Survival Rate of E. coli in Soil (A) and Transition of the Survival Rate of the Wild Type Strain of E. coli in Soil (B)
(2) Identification of novel genes involved in the viability of microorganisms in soil
We next sought to identify genes involved in the soil viability of E. coli using established methods for assessing viability. To achieve this, we focused on a total of about 300 transcription factors in E. coli. Since the expression8 of genes on the genome is regulated by these transcription factors, we hypothesized that analyzing only the transcription factors (rather than the entire genome of approximately 4,400 genes) would allow us to effectively identify the signal transducers9 that microorganisms sense for long-term survival in soil. This approach also aimed to comprehensively clarify the gene functions related to long-term survival. Transcript factors play a crucial role in responding to environmental changes, forming networks that regulate multiple genes based on their roles. Thus, we believed this method would be an effective means to elucidate the singling mechanisms microorganisms use to survive in soil.
We investigated the relationship between each transcription factor and its viability in soil using E. coli strains deficient in individual transcription factor genes. By comparing the viability of these mutants with that of the wild-type strain, we determined whether the transcription factor functions negatively (if the viability of the defective strain increases) or positively (if the viability decreases). Through analysis of 294 defective strains, we identified four strains with increased viability and ten strains with decreased viability due to the deletion of specific transcription factors. Among these, RpoS has been identified as the only transcriptional regulator previously reported to affect long-term soil viability. For the first time, these genes were found to contribute to long-term E. coli survival in soil.
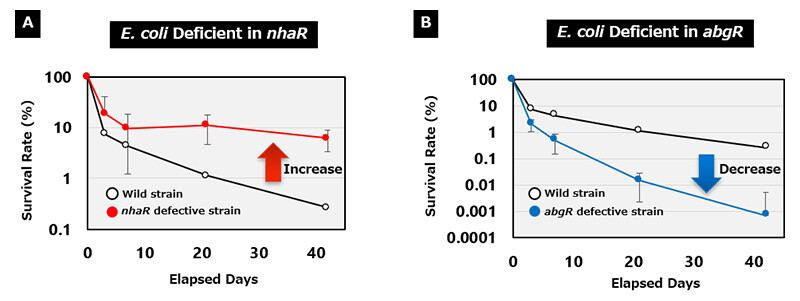 Figure 4 An Example of an Experimental Result of a Transcription Factor That Greatly Affected the Survival Rate of E. coli in Soil.
Figure 4 An Example of an Experimental Result of a Transcription Factor That Greatly Affected the Survival Rate of E. coli in Soil.
(A) Deletion Increases Survival; (B) Deletion Decreases Survival.
(3) Function of the identified gene in the microbe
As mentioned earlier, E. coli is a model microorganism, and extensive knowledge about the function of its genes has been accumulated. Based on previous studies, we summarized the functions of the transcription factors identified in this study. In the analysis, transcription factors whose viability was increased by deletion are marked in red, while those whose viability decreased are marked in blue. These transcription factors were associated with various cellular processes including stationary phase10 stress11 (green), nitrogen source metabolism12 (yellow), carbon source metabolism13 (orange), and osmotic14 stress (blue). These findings suggest that microorganisms utilize genes related to the stationary phase, osmotic stress adaptation, and metabolism of carbon and nitrogen sources to survive in soil for extended periods. The analysis also revealed that these transcription factors are highly conserved among microbial species, indicating their universal role in microorganisms. As described above, this study is the first to clarify the identification and function of transcription factors involved in long-term survival in soil—an aspect that cannot be addressed by short-term analyses using liquid media in laboratory conditions.
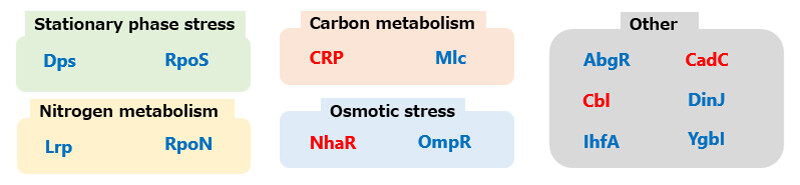 Figure 5 Functional Classification of Identified Genes in Microorganisms
Figure 5 Functional Classification of Identified Genes in Microorganisms
Role of each company
Outlook
This study marks the first comprehensive analysis of all transcription factors in a single bacterium (E. coli) to identify genes essential for long-term survival in soil. Since the identified genes are transcription factors, further investigation into the genes they regulate will enhance our understanding of the molecular mechanisms underpinning soil survival. Additionally, transcription factors are known to alter their function in response to environmental signals, such as nutrient availability. By identifying and leveraging these signals, it may be possible to control viability through gene expression. The application of these findings extends beyond the model organism E. coli to include microorganisms involved in the nitrogen cycle. This could lead to a reduction in greenhouse gas emissions, lower levels of excess nitrogen released into the environment, and minimized environmental impact of chemical fertilizers. For instance, enhancing the survival of microorganisms that convert nitrate to nitrogen and N2O to nitrogen in soil could significantly reduce N2O emissions and excess nitrogen (Figure 6). Given that the soil's nutrient cycle depends heavily on microbial functions, maintaining biodiversity is critical when implementing this technology. Moving forward, we will continue research and development while holistically evaluating the soil's circulation system.
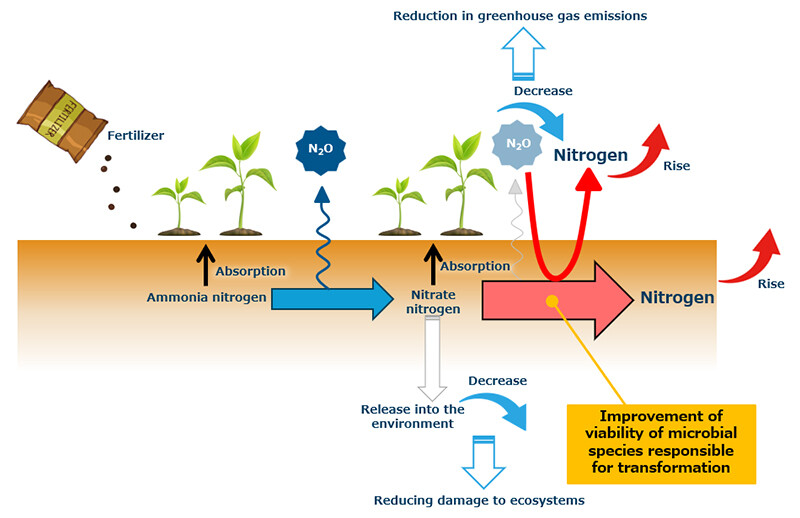 Figure 6 Example of Future Development Using This Fundamental Technology
Figure 6 Example of Future Development Using This Fundamental Technology
[Information on presenters and researchers]
Department of Agricultural Chemistry, School of Agriculture, Meiji University
NTT
Dr. Sousuke Imamura, Senior Distinguished Researcher
[Glossary]
1.Transcription factors: Proteins that specifically bind to DNA and regulate the transcription of specific genes (the step in which DNA is converted to RNA) either by stimulating or inhibiting transcription.
2.Escherichia coli (E. coli): A type of intestinal bacteria, one of the main bacterial species in the human's large intestine, and a common organism in the environment.
3.https://www.env.go.jp/earth/ipcc/6th/index.html(Japanese)
4.The cumulative effect of 1 kg of each gas on global warming over a specified period (e.g.,100 years) compared to that of carbon dioxide (Source: https://www.env.go.jp/policy/hakusyo/h03/7824.html).
5.PCR (Polymerase Chain Reaction): A technique for efficiently amplifying specific DNA fragments.
6.Agar plate: A medium solidified with agar and used to culture and observe microorganisms.
7.Colony: A group of microorganisms growing from a single cell on an agar plate. For example, in the case of E. coli, it forms a circular colony of several millimeters on an agar plate.
8.Gene expression: The process in which information in DNA is converted into functional proteins through transcription (DNA to RNA) and translation (RNA to protein).
9.Signal transducers: Molecules responsible for communication within and between cells that regulate gene expression.
10.Stationary phase: In cell culture and microorganism growth, this is the phase when the number of cells stops increasing and the growth becomes stable.
11.Stress: Refers to the pressure placed on a cell when environmental conditions deviates from optimal. For example, during stationary phase stress, this can occur when stress that inhibits cell growth is applied.
12.Nitrogen metabolism: A series of metabolic processes in which organisms take up nitrogen sources (e.g., ammonia, nitrates, amino acids) from the environment and convert them into nitrogen-containing molecules (e.g., proteins, nucleic acids) required for survival.
13.Carbon metabolism: A series of metabolic processes in which organisms utilize carbon sources (e.g., carbohydrates, fats, proteins) from the environment and use them to convert them into energy and organic molecules (e.g., sugars, lipids, amino acids).
14.Osmotic stress: The force by which water moves from a thinner solution to a thicker solution. It influences water movement inside and outside cells, helping maintain proper cellular balance.
About NTT
NTT contributes to a sustainable society through the power of innovation. We are a leading global technology company providing services to consumers and businesses as a mobile operator, infrastructure, networks, applications, and consulting provider. Our offerings include digital business consulting, managed application services, workplace and cloud solutions, data center and edge computing, all supported by our deep global industry expertise. We are over $93B in revenue and 330,000 employees, with $3.6B in annual R&D investments. Our operations span across 80+ countries and regions, allowing us to serve clients in over 190 of them. We serve over 75% of Fortune Global 100 companies, thousands of other enterprise and government clients and millions of consumers.
Media contacts
NTT Information Network Laboratory Group
Public Relations
Inquiry form
Meiji University
Public Relations Office
Management and Planning Division
03-3296-4082
koho@mics.meiji.ac.jp
<For interview inquiries>
Please fill out this interview application form.
https://www.meiji.ac.jp/koho/purpose/media.html
Information is current as of the date of issue of the individual press release.
Please be advised that information may be outdated after that point.
NTT STORY
WEB media that thinks about the future with NTT